Session Chairs: Michelle West & Doug Lowy
Oral Talk #107
Liang-Wei Wang1,2, Ina Ersing1, Luis Nobre3, Hongying Shen4, Stephen Trudeau1, Camille Ashbaugh1, Steve Gygi5, Vamsi Mootha4, Michael Weekes3, Benjamin Gewurz1,2
1Division of Infectious Disease, Brigham & Women’s Hospital, 2Program in Virology, Harvard Medical School, 3Cambridge Instiute for Medical Research, University of Cambridge, 4Department of Molecular Biology, Massachusetts General Hospital, 5Department of Cell Biology, Harvard Medical School
Epstein-Barr virus (EBV) transforms B-cells into continuously proliferating lymphoblastoid B-cell lines (LCL). Knowledge of how EBV remodels host metabolic pathways to support rapid B-cell outgrowth and to overcome metabolic redox stress remains incomplete. To gain insights into EBV-induced metabolic dependency factors, we constructed a proteomic map of B-cell transformation. Primary B-cells from 12 human donors were profiled by tandem-mass-tag mass spectrometry at rest and at 9 timepoints after EBV infection. This approach generated expression profiles of >6500 host and 17 viral proteins and highlighted EBV-induced pathways. Glycolysis, cholesterol and fatty acid biosynthesis pathways were highly upregulated cytosolic metabolic pathways, each if of which were critical for EBV- driven B-cell outgrowth. Likewise, EBV remodeled B-cell mitochondria, with the one-carbon (1C) metabolism pathway among the most highly EBV-induced. Mitochondrial 1C uses folate carriers to interconvert serine into glycine, formate and NADPH building blocks. EBNA2 and cMyc were critical for upregulation of key mitochondrial 1C enzymes including MTHFD2, which is highly expressed during embryogenesis but not in most adult tissues. Chemical and CRISPR genetic analysis underscored MTHFD2 and 1C pathway roles in EBV-driven B-cell growth and survival. MTHFD2 was important for intramitochondrial NADPH generation, and compartment-specific perturbation of mitochondrial NADPH levels diminished LCL growth and survival. Isotope tracing studies further supported EBV-induced 1C pathway roles in purine nucleotide synthesis and NADPH production. To fuel 1C metabolism, EBV upregulated import and de novo synthesis of serine, each of which were found to be important dependency factors. 1C-derived glycine also supported glutathione synthesis, which together with NADPH had redox defense roles. Our results highlight mitochondrial 1C as a key EBV-induced metabolic dependency factor and potential therapeutic target.
Label with tandem-mass-tag (TMT) based MS3 mass spectrometry
CD23+ of time points 0, 2, 4, 7, 10, 14, 18,21, 28, 35
Day 7, LMP1 and LMP2 —> Burkitt like
Day 14 lymphoblastoid
Latency III antigen expression profiles
See notes for the rest of the talk (extremely fast!)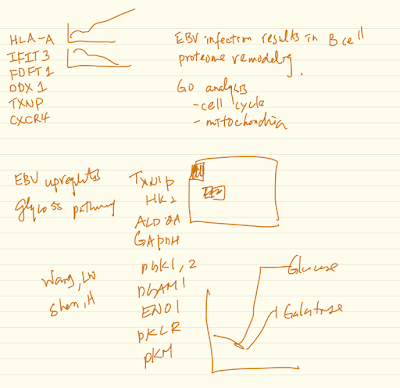
Divya Nandakumar1, Britt Glaunsinger1,2
1University of California Berkeley, 2Howard Hughes Medical Institute
Herpesviruses compete with the tightly regulated cellular transcription by encoding transcription factors that divert the transcriptional machinery from the host to the viral genome. In β and γ-herpesviruses, a viral pre-initiation complex (vPIC) orchestrates the transcription of genes expressed late in infection by hijacking the host RNA polymerase. Identifying candidate genes regulated by the vPIC will be key to detecting promoter elements that are important for vPIC recognition and potentially offer insights on the mechanism of switch from early to late gene expression. In this study, we have used an integrative approach to identify targets of the vPIC using a combination of ChIP-Seq, RNA- Seq and reporter assays. Given that most viral proteins have multiple functions, a targeted approach using site specific mutants in the vPIC that exclusively inhibit transcription would provide more accurate information on vPIC targets. The core component of the vPIC is ORF24, a viral TATA Binding Protein (TBP) that displays putative structural mimicry to cellular TBP but unlike TBP binds directly to both RNA polymerase II and promoter DNA. We have used two specific mutants of ORF24 (a RNA polymerase II interaction mutant and a mutant of ORF24 that disrupts vPIC formation) to identify the functional gene targets of the vPIC. Additionally, we have integrated the differential expression information with genome-wide binding data of the vPIC, generated using an endogenously tagged component of the vPIC, to differentiate the direct and indirect targets of the complex during infection. Our results indicate that the spectrum of genes regulated by the vPIC extends beyond late genes and includes early genes that continue to be expressed at later stages of infection. The results also hint at a link between DNA replication and transcription and current studies are underway to validate the functional significance of the genome-wide analysis.
Six proteins
- ORF24
- ORF31
- ORF34
- ORF18
- ORF30
- ORF66
* A hybrid viral host transcription complex essential for KSHV life cycle (Mol Cell 57, p3490)
* Integrative approach to identify gene targets of KSHV transcription
* ORF24 interacts with RNA Pol II through leucine
* ORF24-Pol II interaction mutant does not produce infectious visions and is detective in alert gene transcription
* Distinct gene clusters that show differential dependence on late gene transcription complex
* ORF24 – dependent
* ChIP-seq of ORF34-HA shows binding of ORF34 across the viral genome
* These represent ORF24 dependent
* Overlap of RNA-seq and ChIP-seq data distinguishes the direct and index
* TATTTAA (direct target short motif)
* Tang et al J Viral 2004 p2609
* K8.1 RSNYS motif
* Expand mitigation us aknost exclusively in direct targets of ORF24 complex
* ORF59 is the only outlier of the
* TATT motif may be important for recognition but downstream sequence sequ is potentially important for transcription
Oral Talk #109
Synthesis
Sebastian O. Wendel1, Laura Brown2, Jazmine Snow1, Tyler Bastian3, Vaibhav Murthy1, Daniel Neill2, Monica Gamez1, Andrew Kahn1, Dalton Dacus1, Kevin Ault4, Ossama Tawfik5, Cen Wu3, Nicholas A. Wallace1
1Division of Biology, Kansas State University, 2Department of Pathology and Laboratory Medicine, University of Kansas Medical Center, 3Department of Statistics, Kansas State University, 4Department of Obstetrics and Gynecology, University of Kansas Medical Center, 5MAWD Pathology Group
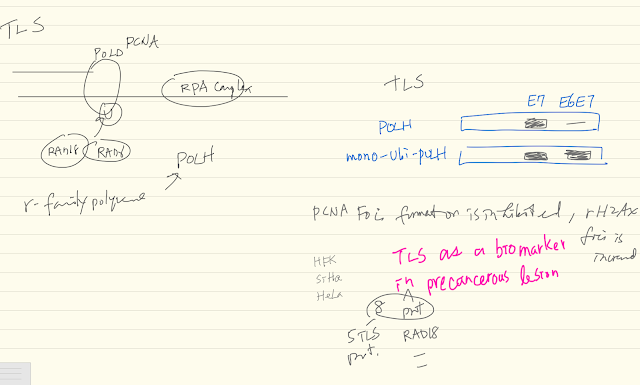
Translesion synthesis (TLS) is an essential DNA-damage tolerance pathway needed during replication. It prevents minor DNA lesions from causing replication fork collapse and descending into more deleterious double strand DNA breaks. Increased proliferation induces TLS to cope with the associated elevation in replication stress. Our bioinformatics analysis shows that increased TLS expression correlates with unfavorable outcomes of patient survival data for several cancer types. Cervical cancers, almost exclusively caused by high risk α-HPV, were the only exception that we found. Here, increased TLS expression is predictive of favorable outcomes. An in-depth bioinformatics analysis of RNAseq data from cervical cancers revealed a general upregulation of the TLS pathway, with the notable exception of POL-η, the lesion bypassing TLS polymerase. HPV E7 driven proliferation causes replication stress. In addition to increased mRNA, a subset of TLS- pathway proteins is stabilized in α-HPV oncogene expressing cervical cancer cell lines. Increases in TLS expression (protein abundance and RNAseq) correlate with disease progression. However, we show that TLS-functionality falls apart in the presence of E6. HPV E6 interferes with the induction of the TLS polymerase POL-η by degrading p53 and blocking its role as transcription factor for POL-η induction. HPV-positive cervical cancer cell lines fail to increase POL-η abundance or concentrate the polymerase in response to extrinsic DNA damage. This increases replication fork stall and collapse, indications of failed TLS. Consequently, we show that cervical cancer cell lines exhibit a significant sensitivity to chemical DNA-crosslinkers as well as UV radiation. We also provide evidence of a subpopulation (~20%) of cervical cancer patients that have increased expression of y-family polymerases. These women have a dramatically decreased survival. Our data makes the case that α-HPV oncogenes E6 and E7 both disrupt and induce the TLS pathway. Further, because TLS mitigates the damage from crosslinking agents, y-family polymerase expression could potentially be used as a diagnostic to identify cervical cancer patients that are unlikely to respond to the standard of care, Cisplatin. Resolving cervical cancer patient data by stage suggests a progression and correlation of RNAseq and immunohistochemistry data with increased cancer stage.
- Hyperproliferation of
- TLS abundance correlated with proliferation
- TLS pathway
- upregulation correlates with survival in cervial cancer
- 5 datasets merged
- 188 subjects
- 5 major TLS related enzymes (POLH, …) are either located at non-changed groups
- POLH abundance in response to UV radiation
- POLH expression correlates with p53 expression
- Nonetheless, About 20% patients with TLS proteins go down associated with poor prognosis
Repair Pathway
Sujita Khanal1, Denise Galloway1
1Division of Human Biology, Fred Hutchinson Cancer Research Center
Persistent expression of high-risk HPV oncogenes is necessary for the development of anogenital and oropharyngeal cancers. Our study showed that E6/E7 expressing cells are hypersensitive to the DNA crosslinking agent cisplatin and have defects in repairing DNA inter strand cross-links (ICL), suggesting that HPV oncogenes impair the Fanconi anemia (FA) pathway. Monoubiquitination and foci formation of FancD2 are functional activators of the FA pathway. Consistent with previous studies, we found that E6/E7 increased FancD2 monoubiquitination and foci formation. However, using an I-SceI co-localization assay revealed that HPV E6 and E7 either expressing individually or together impair the co- localization of FancD2 with double-strand breaks (pH2AX). Recently, de-ubiquitination of FancD2 was recognized to be equally important as ubiquitination for effective ICL repair. We demonstrated that E6 caused delayed FancD2 de-ubiquitination, thereby impairing ICL repair. E6 cells also showed defects in repair proteins, Rad51 and Palb2, which are downstream of FancD2 in the FA pathway. Using genetic epistasis analyses, we showed that mislocalization defect of Rad51 in E6 cells is dependent on FancD2. Further, E6 reduced the expression and foci formation of Palb2. These findings uncover unique mechanisms by which HPV oncogenes (notably E6) impair the completion of the FA repair pathway, though they activate the initiation of the pathway.
Developing Inhibitors Interrupting KSHV Capsid Assembly
Danyang Gong1, Xinghong Dai1, Ting-Ting Wu1, Hong Z. Zhou1, Ren Sun1
1UCLA
Kaposi’s sarcoma-associated herpesvirus (KSHV) capsid, like those of the other eight human herpesviruses, is highly pressurized by its dsDNA genome. Indeed, capsid assembly and genome packaging of herpesviruses are prone to interruption and thus can be targeted for structure-guided antiviral development. However, with nearly 3,000 proteins and over 1,300 Å in diameter, herpesvirus capsids present a formidable challenge to atomic structure determination and functional mapping of molecular interactions. By employing cryo electron microscopy (cryoEM) with highly purified KSHV virions, we resolved a 4.2 Å resolution structure of the KSHV capsid, and its atomic model containing unique conformers of the major capsid protein (MCP), the smallest capsid protein (SCP), the triplex proteins Tri1 and Tri2. Guided by the atomic resolution structure of KSHV in addition to functional analysis results, we have identified interaction hotspots essential for KSHV capsid assembly and replication, especially a groove on the upper domain of MCP enabling its interaction with a long helix on the SCP. More importantly, this groove could be targeted by SCP helix-mimicking polypeptides for inhibiting KSHV replication, making it a promising druggable site for developing inhibitors. Furthermore, the in vitro and in vivo MCP-SCP interaction systems are established and applied to identify chemical inhibitors using high throughput chemical screening.
955 MCP and SCP complex in each capsid
High pressure during DNA packaging