由 ingrid 在 二, 04/01/2014 – 13:03 發表 Cholangiocarcinoma FGFR fusion 高醫 黃道揚
Dear SF,
My mailing address is:
高雄市三民區自由一路100號
5F腎功能室.
高雄市三民區自由一路100號
5F腎功能室.
———————–
SFL1=988 ng/ul C9 RNA 15 ug = 15ul (希望他能訂出FGFR3-TACC3)
SFL2=792 ng/ul SW780 RNA 15 ug = 20ul (希望他能訂出FGFR3-BAIAP2L1)
SFL3=1100 ng/ul TW2.6 RNA 15 ug = 13ul(希望他訂不出任何FGFR3 fusion)
#1由 sufang 在 五, 07/25/2014 – 11:29 發表。
7/24 黃醫師來函
Daw-yang Hwang 黃道揚 to me, 陳Leo
Dear Prof Chen and SF,
Dear Prof Chen and SF,
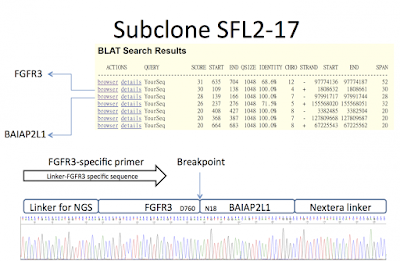
I got the positive result of the SFL2 RNA after some trouble shooting of the experimental designs. Please see the attached file, with FGFR3-BAIAP2L1 fusion sequence. Please let me know if this result is correct? I am waiting for the SFL1 result.
I think if TOPO sucloning can find the fusion products, I believe this method will work better when I put them into MiSeq machine. Potentially, multiple TK detection primers should work better than only one detection primer, since the NGS like more complex heterogeneous fragments as input.
Kindly regards,
Daw-yang
Daw-yang
—————————
Su-Fang Lin to Daw-yang Hwang., 陳Leo, 英潔YCK
Dear Daw-yang,
Here is the answer posted on LINE from my lab associate :
「根據怡宓的論文supplement,序列斷點是一樣的,黃醫師成功了!」
So Congratulations!
I am wondering what made it work this time? Did you change primer locations? And what about bands in SFL3, the sample supposed to be “no fusion” Thanks!
—————————
Daw-yang Hwang 黃道揚 8:57 PM (14 hours ago)
to me, 陳Leo
to me, 陳Leo
I think the main point is the cDNA concentration. The protocol from the company may not be the best.
For the SFL3, I think that is a false positive product, since the FGFR3 specific primer has some overlap with the fusion partner.
For the SFL3, I think that is a false positive product, since the FGFR3 specific primer has some overlap with the fusion partner.
#1 Daw-yang Hwang 黃道揚 8:57 PM (14 hours ago)
to me, 陳Leo
I think the main point is the cDNA concentration. The protocol from the company may not be the best.
For the SFL3, I think that is a false positive product, since the FGFR3 specific primer has some overlap with the fusion partner.
#2由 sufang 在 四, 07/24/2014 – 08:38 發表。 7/17 黃醫師來函
Dear SF,
The initial result of my method showed some positive clones. The SFL3 has might have a FGFR3-AREL1 fusion, but I am not sure it is real or false positive cloning. I am repeating this (SFL3) and other 2 samples.
Furthermore, the FGFR3 primer I designed in at position c.2245-2264 (uniPortKB P22607, a.a. 806). I would like to know that if your known FGFR3 fusion proteins’ breakpoint are down-stream of this region (i.e. after FGFR3 amino acid position 756 )?
I will go to University of Michigan at the end of this month, maybe I can email your friend (YM Wu) first to see if I can pay them a visit?
Kindly regards,
Daw-yang
Daw-yang
—————————-
Su-Fang Lin  Jul 17
Jul 17
 Jul 17
Jul 17Dear Daw-Yang,
怎會降子! 我翻出那時的小抄,是這樣子寫的:
——————————
——————————
SFL1=988 ng/ul C9 RNA 15 ug = 15ul (希望他能訂出FGFR3-TACC3)
SFL2=792 ng/ul SW780 RNA 15 ug = 20ul (希望他能訂出FGFR3-BAIAP2L1)
SFL3=1100 ng/ul TW2.6 RNA 15 ug = 13ul(希望他訂不出任何FGFR3 fusion)
——————————
至於primer位點問題:
C9/SFL1我們有託Dan他們做RNA-seq,
Dr. Yi-Mi Wu那邊我下午再來寫個email介紹你們認識!
Best,
Su-Fang
Su-Fang
#3由 sufang 在 二, 04/01/2014 – 13:18 發表。
另外…
他還希望我們可以給他一份我們常用來轉cDNA的protocol. 我想給他sperscriptIII這個MSDS,然後告訴他通常1 ug起跳,大家覺得這樣子說OK嗎?
#4由 sufang 在 三, 04/02/2014 – 09:47 發表。
email on the next day (4/2/2014)
Dear Daw-Yang,
You will receive three samples, please just throw them into -80C at this moment. Their OD260/280 ratio looked OK and if you need the detailed numbers, I can forward them to you later. Their respective concentrations are:
SFL1=988 ng/ul (15ul in formamide)
SFL2=792 ng/ul (20ul in formamide)
SFL3=1100 ng/ul (13ul in formamide)
SFL2=792 ng/ul (20ul in formamide)
SFL3=1100 ng/ul (13ul in formamide)
We usually take out 1-2 ug RNAs from the formamide stock, make it to be 10 ul solution in clean water, add 1/10 vol salt (1-ul) and 3 vol cold ethanol (30-ul), precipitate in 20C for more than 30 min, spin down and perform cDNA synthesis (please see attached pdf for details).
Please drop me a line once the package arrives safely! Thank you~