由 natsumi 在 四, 07/17/2014 – 17:30 發表 EBV and KSHV Methylation
最近聽Dr. Lin講Methylation的故事,我想到之前曾經做過的一個小實驗,分享之。
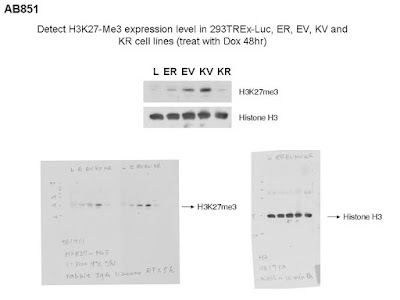
AB851
#1由 ingrid 在 五, 07/18/2014 – 12:20 發表。
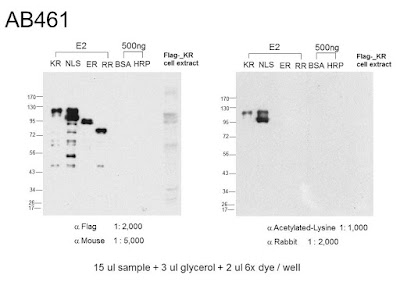
ER沒有被acetylated-lysine Ab認到
#2由 ingrid 在 五, 07/18/2014 – 15:30 發表。
其他Data
#3由 sufang 在 五, 07/18/2014 – 14:48 發表。
是KR!
記錯了,所以ER什麼都沒有! 可能只有phosphorylation.
-而且只有
Ser/Thr-phosphorylation.
#4由 sufang 在 五, 07/18/2014 – 09:10 發表。
H3K27me3是Histone這邊的repressive mark
有它在的地方,基因多為repressive (for transcription)。這是要說lytic cycle時EBV及KSHV設法把H3K27me3的histone穩定住(via ubiquitination system?),並且加在viral genome上,讓病毒下一個round一進入宿主細胞,大部份基因不表現?
小結一下:
Repressive: H3K27me3 / DNA methyltion low
Active: H3K4me3
Enhancer: H3K4me1
Elongation: H3K36me3 / DNA methylation high
另外:
CBX5=HP1
CBX1=HP1-BETA
CBX3=HP1-GAMMA
(怎麼記? 摔筆、翻桌#$&*^%$!+_=*^)
#5由 sufang 在 五, 07/18/2014 – 08:39 發表。
兩篇很好的文章分享
Nat Rev Cancer. 2011 Sep 23;11(10):726-34. doi: 10.1038/nrc3130.
A decade of exploring the cancer epigenome – biological and translational implications.
Baylin SB1, Jones PA.
Abstract
The past decade has highlighted the central role of epigenetic processes in cancer causation, progression and treatment. Next-generation sequencing is providing a window for visualizing the human epigenome and how it is altered in cancer. This view provides many surprises, including linking epigenetic abnormalities to mutations in genes that control DNA methylation, the packaging and the function of DNA in chromatin, and metabolism. Epigenetic alterations are leading candidates for the development of specific markers for cancer detection, diagnosis and prognosis. The enzymatic processes that control the epigenome present new opportunities for deriving therapeutic strategies designed to reverse transcriptional abnormalities that are inherent to the cancer epigenome. PMID: 21941284
Genome Res. 2014 Feb;24(2):177-84. doi: 10.1101/gr.157743.113. Epub 2013 Sep 25.
Large-scale hypomethylated blocks associated with Epstein-Barr virus-induced B-cell immortalization.
Hansen KD1, Sabunciyan S, Langmead B, Nagy N, Curley R, Klein G, Klein E, Salamon D, Feinberg AP.
Author information
Abstract
Altered DNA methylation occurs ubiquitously in human cancer from the earliest measurable stages. A cogent approach to understanding the mechanism and timing of altered DNA methylation is to analyze it in the context of carcinogenesis by a defined agent. Epstein-Barr virus (EBV) is a human oncogenic herpesvirus associated with lymphoma and nasopharyngeal carcinoma, but also used commonly in the laboratory to immortalize human B-cells in culture. Here we have performed whole-genome bisulfite sequencing of normal B-cells, activated B-cells, and EBV-immortalized B-cells from the same three individuals, in order to identify the impact of transformation on the methylome. Surprisingly, large-scale hypomethylated blocks comprising two-thirds of the genome were induced by EBV immortalization but not by B-cell activation per se. These regions largely corresponded to hypomethylated blocks that we have observed in human cancer, and they were associated with gene-expression hypervariability, similar to human cancer, and consistent with a model of epigenomic change promoting tumor cell heterogeneity. We also describe small-scale changes in DNA methylation near CpG islands. These results suggest that methylation disruption is an early and critical step in malignant transformation. PMID: 24068705