#1由 sufang 在 一, 05/18/2015 – 08:50 發表。
Keratinocyte culture method
說在前面:
Rheinwald Lab所寄給我們的Human Keratinocyte Culture Methods第一段便比較下面三個廠商來的media, 說請大家用K-sfm來養”low-to-moderate density”的keratinocytes (primary or immortalized)
| K-sfm | Life Technologies | grow well from low to moderate density |
| EpiLife | Life Technologies | |
| KGM (or MCDB154) | Clonetics | very short replicative lifespan |
第二段:
須牢記於心的是, K-sfm的營養成分僅適用於低密度、聚集性的keratinocyte生長, 因此用K-sfm養的細胞, 1/3滿度前就必須passage.
第三段: 碰到OSCC的話, 建議與irradiated 3T3 fibroblasts共養、養在DMEM/F12 + 10-20% calf serum + 0.4 ug/ml hydrocortisone.
以下是養細胞步驟, Adapt from 宛樺labnotes and Rheinwald Lab.
For a p100 dish (or T75) –> plaing 104 – 105 cells / dish
For a p60 dish (or T25) –> plaing 103 – 104 cells / dish
Cultures are refed every two days.
5-8 days after plating, cells are ready for the next passage.
–It is important to subculture keratinocytes no more than 8 days after they have been plated, even if they have not reached the desired density (每一繼代不超過8天).
–A healthy, proliferative cell population should contain primarily small cells which have a population doubling time of ~24-30 hours (正常健康的小細胞約24–30小時複製一代)
–It is best practice to count the cells at each subculture and replate at a defined number, avoid the common practice of many labs for serially passaging cell lines of plating 1:4 or 1:10 dilutions (維持每次繼代種入固定細胞數, 不要用1:4, 1:10這種不良繼代習慣).
Growing keratinocytes to high density (提高細胞培養密度之培養方法)
–K-sfm medium was not designed to grow cells to high density with retention of viability. To grow keratinocytes to high density a medium containing higher concentrations of nutrients is needed. We have found that keratinocytes and SCC cells that have been grown up to ~1/3 confluence in K-sfm can be switched at this time to a 1:1 mixture of K-sfm and a medium we call “DF-K” (note: K-sfm + DF-K –> KDF medium).
–K-sfm medium was not designed to grow cells to high density with retention of viability. To grow keratinocytes to high density a medium containing higher concentrations of nutrients is needed. We have found that keratinocytes and SCC cells that have been grown up to ~1/3 confluence in K-sfm can be switched at this time to a 1:1 mixture of K-sfm and a medium we call “DF-K” (note: K-sfm + DF-K –> KDF medium).
–SF notes: 請大家漸漸把immortalized keratinocytes 馴服在KDF medium中, 謝謝! These incudes
* CGHNK2
* OKF4hTERT
* NP460hTERT
* NP550hTERT
* DOK
Recipe for the above Rheinwald lab modification of KSFM:
(GIBCO K-sfm +25 µg/ml BPE +0.4 mM CaCl2 +0.2 ng/ml EGF +pen/strep)
(GIBCO K-sfm +25 µg/ml BPE +0.4 mM CaCl2 +0.2 ng/ml EGF +pen/strep)
500 ml K-sfm (Invitrogen/Life Technologies, which comes with 0.1 mM CaCl2)
½ vial Life Technologies’ tube containing 25 μg Bovine Pituitary Extract (BPE)
0.5 ml 0.3 M CaCl2 in water (=1000x for 0.3mM)
0.5 ml 0.2 μg/ml EGF in PBS +0.1%BSA (=1000x for 0.2 ng/ml)
5 ml Penicillin/Streptomycin (100X conc. solution) (Life Technologies)
Swirl to mix and then filter-sterilize through a 0.2 μm pore-size sterilization filter. The medium remains good for at least one month at 4oC.
½ vial Life Technologies’ tube containing 25 μg Bovine Pituitary Extract (BPE)
0.5 ml 0.3 M CaCl2 in water (=1000x for 0.3mM)
0.5 ml 0.2 μg/ml EGF in PBS +0.1%BSA (=1000x for 0.2 ng/ml)
5 ml Penicillin/Streptomycin (100X conc. solution) (Life Technologies)
Swirl to mix and then filter-sterilize through a 0.2 μm pore-size sterilization filter. The medium remains good for at least one month at 4oC.
Recipe for DF-K medium:
(DMEM + F12 (1:1 v/v) +2 mM L-glutamine +25 µg/ml BPE +0.4 mM CaCl2 + 0.2ng/ml EGF )
(DMEM + F12 (1:1 v/v) +2 mM L-glutamine +25 µg/ml BPE +0.4 mM CaCl2 + 0.2ng/ml EGF )
250 ml DMEM (High Glucose, no pyruvate, no glutamine, no calcium (Life Technologies catalog #21068-028)
250 ml Ham’s F-12 medium (Life Technologies catalog #11765-054)
3.75 ml 200 mM L-Glutamine (=100x for 2 mM) (Invitrogen/Life Technologies)
½ vial Life Technologies’ tube containing 25 μg Bovine Pituitary Extract (BPE)
0.42 ml 0.3 mM CaCl2 in water (=1000x for 0.3mM)
0.5 ml 0.2 μg/ml EGF in PBS +0.1%BSA (=1000x for 0.2 ng/ml)
250 ml Ham’s F-12 medium (Life Technologies catalog #11765-054)
3.75 ml 200 mM L-Glutamine (=100x for 2 mM) (Invitrogen/Life Technologies)
½ vial Life Technologies’ tube containing 25 μg Bovine Pituitary Extract (BPE)
0.42 ml 0.3 mM CaCl2 in water (=1000x for 0.3mM)
0.5 ml 0.2 μg/ml EGF in PBS +0.1%BSA (=1000x for 0.2 ng/ml)
5 ml Penicillin/Streptomycin (100X conc. solution) (Life Technologies)
Once cultures have grown to ~1/3 confluence in KSFM they either should be subcultured or else subsequently fed daily with a 1:1 mixture of KSFM and DF-K until confluent. Such cultures are suitable for protein, RNA, and DNA extracts or FACS analysis, but may not subculture as well as cells grown to no more than ~1/3 confluence in K-sfm.
#2由 EVKVLIN 在 二, 05/12/2015 – 15:58 發表。
2014/08/15 統計相關
Let’s give statistics the attention it deserves (2013-Aug-29)
Nature Methods | Methagora
Let’s give statistics the attention it deserves
This month we launch a new column ‘Points of Significance’ devoted to statistics, a topic of profound importance for biological research, but one that often doesn’t receive the attention it deserves.
For the past three years Nature Methods has been publishing the Points of View column, one page a month dedicated to practical advice for researchers on how to create accessible and accurate visualizations of their data. The response to the column articles has been fantastic and most recently we organized them by topic here on our blog.
Unfortunately, a truth about data visualization is that no matter how good the visualization, if the experiment wasn’t appropriately designed and the data wasn’t analyzed correctly, the resulting visual depiction of the data will be inherently flawed. Nature Methods and the other Nature journals recently made changes to improve data and methods reporting as part of a reproducibility initiative. We feel this is an important first step in improving experimental reproducibility and repeatability, but unfortunately by the time work is submitted for publication it can be difficult to correct shortcomings in experiemntal design and analysis.
Update: Below is a continuously updated list of the Points of Significance articles.
Importance of being uncertain – September 2013
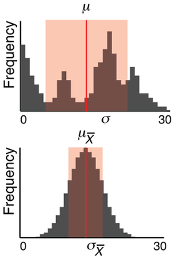
How samples are used to estimate population statistics and what this means in terms of uncertainty.
Error Bars – October 2013
The use of error bars to represent uncertainty and advice on how to interpret them.
Significance, P values and t-tests – November 2013
Introduction to the concept of statistical significance and the one-sample t-test.
Power and sample size – December 2013
Using statistical power to optimize study design and sample numbers.
Visualizing samples with box plots – February 2014
Introduction to box plots and their use to illustrate the spread and differences of samples.
Comparing samples—part I – March 2014
How to use the two-sample t-test to compare either uncorrelated or correlated samples.
Comparing samples—part II – April 2014
Adjustment and reinterpretation of P values when large numbers of tests are performed.
Nonparametric tests – May 2014
Use of nonparametric tests to robustly compare skewed or ranked data.
Designing comparative experiments – June 2014
The first of a series of columns that tackle experimental design shows how a paired design achieves sensitivity and specificity requirements despite biological and technical variability.
Analysis of variance and blocking – July 2014 (Free Access during July)
Introduction to ANOVA and the importance of blocking in good experimental design to mitigate experimental error and the impact of factors not under study.
How samples are used to estimate population statistics and what this means in terms of uncertainty.
Error Bars – October 2013
The use of error bars to represent uncertainty and advice on how to interpret them.
Significance, P values and t-tests – November 2013
Introduction to the concept of statistical significance and the one-sample t-test.
Power and sample size – December 2013
Using statistical power to optimize study design and sample numbers.
Visualizing samples with box plots – February 2014
Introduction to box plots and their use to illustrate the spread and differences of samples.
Comparing samples—part I – March 2014
How to use the two-sample t-test to compare either uncorrelated or correlated samples.
Comparing samples—part II – April 2014
Adjustment and reinterpretation of P values when large numbers of tests are performed.
Nonparametric tests – May 2014
Use of nonparametric tests to robustly compare skewed or ranked data.
Designing comparative experiments – June 2014
The first of a series of columns that tackle experimental design shows how a paired design achieves sensitivity and specificity requirements despite biological and technical variability.
Analysis of variance and blocking – July 2014 (Free Access during July)
Introduction to ANOVA and the importance of blocking in good experimental design to mitigate experimental error and the impact of factors not under study.
_________________
Human Buccal Cell DNA Isolation With a Qiagen QIAamp Mini Kit
(/lims/files/users/sufang/Protocols/human_buccal_cell_dna_isolation_with_a_qiagen_qiaamp_mini_kit.pdf)
QIAamp DNA Blood_Isolation of genomic DNA from saliva and mouthwash
(/lims/files/users/sufang/Protocols/qiaamp_dna_blood_isolation_of_genomic_dna_from_saliva_and_mouthwash.pdf)
QIAamp DNA Mini and QIAamp DNA Blood
(/lims/files/users/sufang/Protocols/qiaamp_dna_mini_and_qiaamp_dna_blood.pdf)
Isolation of DNA from buccal cells using the EZ1 DNA Tissue Kit
(/lims/files/users/sufang/Protocols/isolation_of_dna_from_buccal_cells_using_the_ez1_dna_tissue_kit.pdf)
陽明張國威教授已開放之學生論文電子檔
2004_07_羅尹伸_染色體 11q13
(/lims/files/users/sufang/Unclassidied/2004_07_luo_yin_shen__ran_se_ti__11q13.pdf)
2004_07_張筱清_染色體 3q26-27
(/lims/files/users/sufang/Unclassidied/2004_07_zhang_xiao_qing__ran_se_ti__3q26-27.pdf)