General
Fluidigm Access Array + MySeq
–YouTube 1: https://www.youtube.com/
–YouTube 2: https://www.youtube.com/
–AA_Illumina_ug_100-3770 (pdf)
台大郭頌鑫醫師抽取DNA and RNA from tiny endoscopic biopsy samples使用。
–陳華鍵老師他們新開的公司行動基因,用的是類似方法、只是不同機器(Ion Torrent PGM/Proton)
–Archer: Archer™ FusionPlex™ FGFR Panel (USD 495 / 8 rxn)
–與台大合作部分,詳見HPV and p16 in HNSCC. (按我連結)
–ds-cDNA protocal from Yi-Mi and Dan (03/05/2015). (按我下載)
–ds-cDNA protocal from 黃醫師Lab 智娟 (2015/04/19). (按我下載)
Gene Fusion Panel
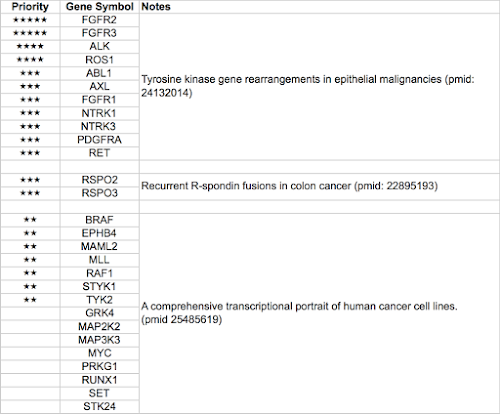
ALK, AXL, FGFR1, FGFR2, FGFR3, NTRK1, NTRK3, PDGFRA, RET, ROS1, MUSK, EGFR,INSR,INSRR, MET, NTRK2, RSPO2, RSPO3
–黃醫師 final list on 2015/04/10
ALK, ABL1, FGFR1, FGFR2, FGFR3, NTRK1, NTRK3, PDGFRA, PDGFRB, RET, ROS1, FLT3
(4 x 96 primers)
Cholangiocarcinoma Panel
ARAF, ARID1A, ARID2, AXIN1, BAP1, BRAF, CDH1, CDKN2A, CTNNB1, FBXW7, FLT4, IDH1, IDH2, KRAS, MLL3, NRAS, PBRM1, PI3K3CA, PTEN, SMAD4, TERT, TP53, VEGFC, VEGFR3,
hTERTp -124, and hTERTp -126 (526 amplicons)
2015/04/20 Line messages from Dr. Huang: (1) 50 ng for one panel, 2 panels for each TLCN sample. (2) 2 PCRs to increase specificity (3) if possible, ship 500 ng / 10 ul / sample will be the best.
TK Panel
Dear Prof Lou,
We will order your primer tomorrow. Beside those oral pharyngeal CA panel, we already have some TK panel, which include: FGFR1, FGFR2, FGFR3, ALK, ROS1, NTRK1, NTRK3, RET, PDGFRA, PDGFRB, FLT3. More TK genes will be added later. We can run your samples in your cancer panel and current TK panel if you are interested. (2015/04/29 email)
#1由 sufang 在 一, 07/20/2015 – 09:37 發表。
2015/07/15 TLCN案號94聯絡窗口
Su-Fang Lin <sflin1@gmail.com> 2015/07/20 (Mon)
to 陳所長, 姜乃榕醫師, Daw-yang Hwang., Evita, 劉奕宏, bcc: 玉蓮YLC
檢體真的快到手了! 謝謝大家的幫忙~ Regards, Su-Fang
———- Forwarded message ———-
From: 陳玉蓮 <cyl@nhri.org.tw>
Date: 2015-07-15 10:58 GMT+08:00
Subject: RE: TLCN案號94 聯絡窗口
To: Su-Fang Lin <sflin1@gmail.com>
Dear Dr Lin
剛剛雅婷有打來說: 下星期她們準備好的 我們可以領到的檢體是90例DNA,43例有T/N的RNA和2例只有T的RNA
RNA他說是目前有的,之後會再補5例T/N或3例T/N (之後補的RNA其對應之DNA也是會一併給 就是3或5例 看之後黃醫師怎麼說),我問了剩下的RNA會再等多久? 可以的話我們希望一起領… 雅婷是說還不知道要看黃醫師,有可能會要一陣子…
那我們是要回覆等檢體齊了再一併拿嗎? 還是就先領來?
另外空白切片正在切,也是之後補。
目前檢體是在-80度西保存,運送的部分,我們自取,她們附上的乾冰額外要500元。
請問TLCN申請的帳 要報哪個計畫? 因為取檢體前要先開發票,所以確定後我再回覆雅婷,費用是23345+500=23845。
玉蓮
2015/07/17 by LINE message
玉蓮: 雅婷說 下週會有48例 T/N RNA 唷 已準備好了
不知道為什麼突然sample都好了^^
費用變成 24195.
陳玉蓮 to me, mmiiee313, haloedition8, 柯英潔 2015/07/20 10:40 am
Dear Dr Lin:
目前知道的資訊
1. 流程: 等發票開好了,她們會通知我們,我們就可以去領檢體了。(這周會開發票)
2. Sample 的濃度: 5ug (大概 500-1000ng/ul) 裝在1.5-ml Eppendrof。
3. 到時我們會領到:
DNA (180管)==> 在3盒小白盒,她們習慣每80管放一盒。
RNA (96管)==> 在2盒小白盒
濃度表(紙本),有要求領sample當天也寄一份電子檔給我們。
發票與1估價單。
PS:
需要先訂好乾冰。
可能要拿兩個略大的保麗龍,推推車上去拿較方便。
10 ul-filter Tip陳家之前買很多,聖萍學姐說可以給我們用。
玉蓮
_______
#2由 sufang 在 五, 02/13/2015 – 16:12 發表。
2015/02/01 Our IHCC project from NJ
姜乃榕 <njchiang@nhri.org.tw> Sun, Feb 1, 2015 at 12:14 AM
To: SuFang Lin <sflin1@gmail.com>, 黃道揚醫師 <910208@mail.kmuh.org.tw> Cc: 陳立宗 <leochen@nhri.org.tw>
Dear All: 大家覺得這篇paper跟我們即將要做的是不是很像? for your referecne!
Best
NJ
Mutation profiling in cholangiocarcinoma prognostic and theraeputic impliccaions.pdf 1621K
********
SuFang Lin <sflin1@gmail.com> Sun, Feb 1, 2015 at 10:58 AM
To: 姜乃榕 <njchiang@nhri.org.tw>
Cc: 黃道揚醫師 <910208@mail.kmuh.org.tw>, 陳立宗 <leochen@nhri.org.tw>
很像, but guess what, I think it is even more similar to what 閻紫宸教授與行動 基因合作的東西 (45 core genes on oral cancer FFPE samples using ion torrent PGM).行動基因陳華鍵、陳淑貞博士2/9星期一會到竹南給演講,到時候我會好 好核對一下 (gene list).
做這個東西 clinical sample 的品質很重要.這篇paper是20% tumor cellularity, 連切十片FFPE, 抽取tumor part的DNA, 10ng as starting material.
黃醫師TLCN那邊的DNA material是由tissue而來, 她說tumor cellularity > 50%. 基因list也要猜得smart. 大家加油!
SuFang
********
Dawyang Hwang <910208@kmuh.org.tw> Wed, Feb 4, 2015 at 8:34 AM
To: 姜乃榕 <njchiang@nhri.org.tw>, SuFang Lin <sflin1@gmail.com> Cc: 陳立宗 <leochen@nhri.org.tw>
Dear Drs. Chiang, Lin, and Prof Chen,
It is nice to meet you this Monday and wish my talk is not too bad.
I wonder if you can arrange the priority gene list of cholangiocarcinoma, and I will calculate the total number of primers and order them before Chinese New year. I plan finish the cholangio panel no late than April if we get the TLCN sample very soon.
Sincerely, Dawyang
********
陳立宗 <leochen@nhri.org.tw>
To: Dawyang Hwang <910208@kmuh.org.tw>, 姜乃榕 <njchiang@nhri.org.tw>, SuFang Lin <sflin1@gmail.com>
Dear DY:
You talk was great, looking forward to having productive collaboration with you! Best regards,
LTC
********
姜乃榕 <njchiang@nhri.org.tw>
To: 陳立宗 <leochen@nhri.org.tw>, Dawyang Hwang <910208@kmuh.org.tw>, SuFang Lin <sflin1@gmail.com>
Dear Dr. Hwang:
It’s so nice for your talk. I’ll make the gene lists ASAP. By the way, I am going to apply the fresh frozen tissue of IHCC from NCKU hospital. Hope we’ll have more dataset.
Thanks again.
Best
********
Su-Fang Lin <sflin1@gmail.com>
To: 姜乃榕 <njchiang@nhri.org.tw>
Cc: 陳立宗 <leochen@nhri.org.tw>, Dawyang Hwang <910208@kmuh.org.tw>
RIKEN有新paper上架! 天意嗎??
Wholegenome mutational landscape of liver cancers displaying biliary
phenotype reveals hepatitis impact and molecular diversity
SF
********
SuFang Lin <sflin1@gmail.com> Sat, Feb 7, 2015 at 8:34 PM To: Dawyang Hwang <910208@kmuh.org.tw>
Cc: 姜乃榕 <njchiang@nhri.org.tw>, 陳立宗 <leochen@nhri.org.tw>
Dear Daw-yang: here comes my wishing list!
(1) For the Gene Fusion project
(2) For the ICC mutation project
I also attach an excel file for your reference!
謝謝您!! 素芳合十
********
陳立宗 <leochen@nhri.org.tw> Sat, Feb 7, 2015 at 10:51 PM
To: SuFang Lin <sflin1@gmail.com>, Dawyang Hwang <910208@kmuh.org.tw> Cc: 姜乃榕 <njchiang@nhri.org.tw>
Dear ALL:
We need to discuss the list further!
Best
LT Chen
********
Dawyang Hwang <910208@kmuh.org.tw>
To: 陳立宗 <leochen@nhri.org.tw>, SuFang Lin <sflin1@gmail.com> Cc: 姜乃榕 <njchiang@nhri.org.tw>
No problem, I think Prof. Chen, Dr. Jiang, and Sufang can discuss more, and I will design calculate the number of primers needed.
Dawyang
________
#3由 sufang 在 三, 01/21/2015 – 23:19 發表。
2014/12/05 List made by Daw-yang
Daw-yang Hwang <910208@mail.kmuh.org.tw> Fri, Dec 5, 2014 at 12:07 PM
Dear all,
I made a list of all candidate genes of cholangiocarcinoma, please let me know if I miss any!
GENE BAP1 ARID1A PBRM1 IDH1 IDH2 FGFR2 PIK3CA PTEN PIK3C2G PIK3C2A TP53 CDK2NA KRAS NRAS TGFBR2 SMAD4 CTNNB1 BRCA1 BRCA2 MLL3 GNAS RNF43 MCL1 ERBB3 MET NF1 PTCH1 TSC1 TSC2 MSH2 MSH6 EPHB1 EPHA7 CDH1 INHBA FGFR1 FGFR3
Daw-yang
************
姜乃榕<njchiang@nhri.org.tw> To: Daw-yang Hwang <910208@mail.kmuh.org.tw>, Su-Fang Lin <sflin1@gmail.com>, <k153chen@gmail.com>
Dear Dr. Huang:
There are two additional gene, “ROS1 and ALK”, please. Thanks a lot
Best,
************
Su-Fang Lin <sflin1@gmail.com> Sat, Dec 6, 2014
To: <njchiang@nhri.org.tw>
Cc: Daw-yang Hwang <910208@mail.kmuh.org.tw>, k153chen@gmail.com>
Dear Daw-Yang,
If there are still room for potential gene fusion candidates, RSPO2 and RSPO3, that have been described in colon cancers, seem to be good targets to try, too.
Thank you very much!
Su-Fang