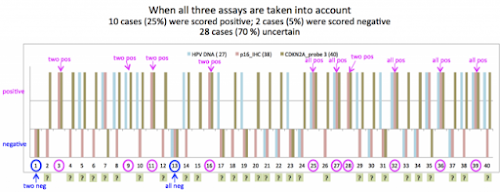
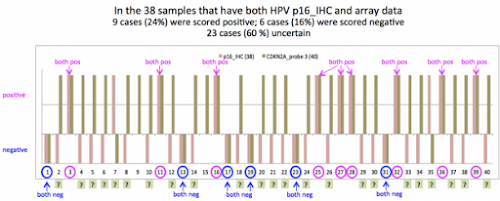
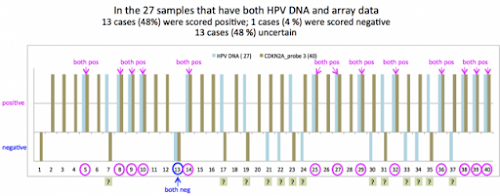
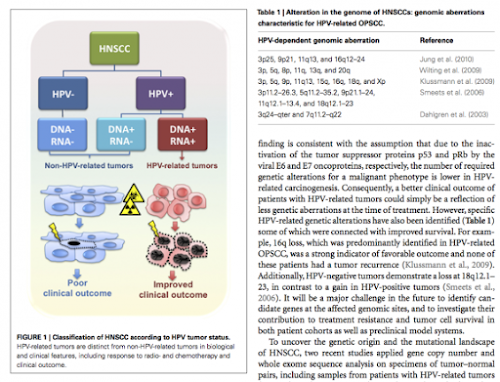
#2由 sufang 在 三, 05/13/2015 – 13:04 發表。
2015/04/29 Re: Gene list/amplicons number for NTU_ENT_ORAL_CA
Dawyang Hwang <darwinhwang@yahoo.com> Wed, Apr 29, 2015 at 7:33 PM ReplyTo: Dawyang Hwang <darwinhwang@yahoo.com>
To: “PeiJen, Alex, Lou” <pjlou@ntu.edu.tw>, “wangcp@ntu.edu.tw” <wangcp@ntu.edu.tw>, “kmu8501094@yahoo.com.tw” <kmu8501094@yahoo.com.tw>, “mayandjean@gmail.com” <mayandjean@gmail.com>
Cc: 李奕錡 <chi171738@gmail.com>, Evita Yu <yu.evita@gmail.com>, SuFang Lin <sflin1@gmail.com>

Dear Prof Lou,
We will order your primer tomorrow. Beside those oral pharyngeal CA panel, we already have some TK panel, which include: FGFR1, FGFR2, FGFR3, ALK, ROS1, NTRK1, NTRK3, RET, PDGFRA, PDGFRB, FLT3. More TK genes will be added later. We can run your samples in your cancer panel and current TK panel if you are interested.
The process will be:
1. Mixing master Primers (we will take <5ul for master Fluidigm mixture, total is about 120ul), we will return your primers when the project is finished.
2. Run Fluidigm chip (access Array)
3. Barcode PCR
4. Purification (步驟14, 48個samples/Chip, cost: 4萬, (1個sample 800元))
5. Run Miseq 2×150 kit, V3 (這一部份外送生技公司 COST比照中研院約 6萬會請生技公司直接向 CHRISTY請款)
6. Data analysis (free for your lab)
Please let me know if there is a problem. Dawyang
********************
Dear Dr Huang,
1. Please contact Ms李奕錡 ( 李奕錡 christy
電話:0233668708 手機: 0927837157
<chi171738@gmail.com> for billing.
2. Yes, you may delete MLL2
3. We will send 47 patients’ DNA and 3 cell lines’ DNA (total 50 samples) to you as the first round testing.
All the best,
PeiJen, Alex, Lou MD, PhD
********************
從: Dawyang Hwang [darwinhwang@yahoo.com]
寄件日期: 2015年4月25日 上午 11:44
至: PeiJen, Alex, Lou; wangcp@ntu.edu.tw; kmu8501094@yahoo.com.tw; mayandjean@gmail.com
主旨: Gene list/amplicons number for NTU_ENT_ORAL_CA
PS: I am sending from yahoo because my email was returned from NTU email system.
Dear Drs. and Prof. Lou:
My lab finished all necessary designs, total amplicon numbers is 689. This will be more than 14x multiplex PCR. I will suggest omit 12 genes (MLL2?) to make them around 12x to reduce failed PCR.
I can order primers and usually arrive after one week.
Kindly regards, Dawyang
PS: please let me know who should we contact about the billing process.
Dear Dr. Wang:
The standard protocol is 1ul with concentration of 50ng/ul. But please give me more (DNA samples) to manipulate since 1ul is will vaporized during shipment. Do you want to give me 48 samples? or you want to put a cellline DNA in them?
Please give me at least 5 ul, best put all samples in a 96well plate (prefer loading in A1H1 then A2 H2, …,A6H6).
You canask you assistant to call me at 0975356113 or 0932351464, for wasy communication. PS: if there is no extra genes, I will order the latest version of the primer design. Regards, Dawyang On Wed, 22 Apr 2015 15:15:35 +0800, chengping wang2 wrote > Hi, Dawyang, > > I am ready to take the DNA of 47 samples to you. Should I take exact 50 ng from each sample to you to do the exp or I can take different amounts, but all more than 50 ng from these 47 samples to you , because I have different DNA concentrations of all specimen in my hand. Which one is convinient to you? >
> 王成平
> ChengPing Wang, M.D.
> Clinical Associate Professor
> Department of Otolaryngology
> National Taiwan University Hospital
> 7 ChungShan South Road
> Taipei, Taiwan
> <wangcp@ntu.edu.tw>
> Tel: (+886)223123456(+886)223123456 ext 63525 > Fax: (+886)223410905
************************
chengping wang2 <wangcp@ntu.edu.tw> Wed, Apr 29, 2015 at 9:23 PM To: Dawyang Hwang <darwinhwang@yahoo.com>
Cc: “PeiJen, Alex, Lou” <pjlou@ntu.edu.tw>, “kmu8501094@yahoo.com.tw” <kmu8501094@yahoo.com.tw>, “mayandjean@gmail.com” <mayandjean@gmail.com>, 李奕錡 <chi171738@gmail.com>, Evita Yu <yu.evita@gmail.com>, SuFang Lin <sflin1@gmail.com>
cheers!
王成平
ChengPing Wang, M.D.
Clinical Associate Professor Department of Otolaryngology National Taiwan University Hospital 7 ChungShan South Road
Taipei, Taiwan
Tel: (+886)223123456 ext 63525 Fax: (+886)223410905
*************************************
PeiJen, Alex, Lou <pjlou@ntu.edu.tw>
To: Dawyang Hwang <darwinhwang@yahoo.com>, “wangcp@ntu.edu.tw” <wangcp@ntu.edu.tw>, “kmu8501094@yahoo.com.tw” <kmu8501094@yahoo.com.tw>, “mayandjean@gmail.com” <mayandjean@gmail.com>
Cc: 李奕錡 <chi171738@gmail.com>, Evita Yu <yu.evita@gmail.com>, SuFang Lin <sflin1@gmail.com>
Dear Dr Huang,
So if we run the cancer panel, how much will be the average cost for 1 sample? And if we run both the cancer and TK panels, how much will be the average cost per sample?
PeiJen, Alex, Lou MD, PhD
Professor
Department of Otolaryngology
National Taiwan University Hospital and National Taiwan University College of Medicine
____________
#3由 sufang 在 三, 05/13/2015 – 12:46 發表。
2015/04/13 Gene list/amplicons number for NTU_ENT_ORAL_CA
Dawyang Hwang <910208@kmuh.org.tw> Mon, Apr 13, 2015 at 4:28 PM To: “PeiJen, Alex, Lou” <pjlou@ntu.edu.tw>, ‘王成平 <wangcp@ntu.edu.tw>, 陳贈成 <kmu8501094@yahoo.com.tw>, 林玫君 <mayandjean@gmail.com>
Cc: SuFang Lin <sflin1@gmail.com>
Dear All,
Please see the attached file for for the oral pharyngeal cancer primers. Few primers need some modification, but I can order them within this week. Since the DNA samples are from FFPE, the primers is designed smaller (less than 200bp), and the amplicon numbers will be around 500 in total 910x11x multiple PCR). Please let me know if you would like to add or delete any genes before I oder tham.
Kindly regards, Dawyang
NTUH_ENT_ORAL_PRIMERS.xlsx 14K
***********
Su-Fang Lin <sflin1@gmail.com>
To: Dawyang Hwang <910208@kmuh.org.tw>
Cc: “PeiJen, Alex, Lou” <pjlou@ntu.edu.tw>, ‘王成平 <wangcp@ntu.edu.tw>, 陳贈成 <kmu8501094@yahoo.com.tw>, 林玫君 <mayandjean@gmail.com>
Thank you very much, Dr. Hwang!
I have just two notes on this primer list:
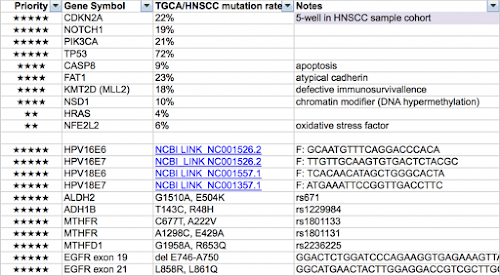
(1) Remember to spare more wells for CDKN2A, after all, it is the main thing for this project.
(2) I thought there are HPV strains, other than HPV16 and HPV18, were detected in these 300 samples by 金車晶片. Should we also consider couple wells for those strains? (如果要刪的話, MLL2是個好選項. 它…實在太大了! 對了, GATA3的status是…?)
SuFang
***********
Dawyang Hwang <910208@kmuh.org.tw> Thu, Apr 16, 2015 at 9:32 PM To: SuFang Lin <sflin1@gmail.com>
Cc: “PeiJen, Alex, Lou” <pjlou@ntu.edu.tw>, ‘王成平 <wangcp@ntu.edu.tw>, 陳贈成 <kmu8501094@yahoo.com.tw>, 林玫君 <mayandjean@gmail.com>
Thu, Apr 16, 2015 at 8:54 PM
Dear All:
We can certainly exam HPV strains (validate Kingcar result?), but I am not familiar with their genotypes, thus someone need to 1. decide which strains to be exam, and 2. find which region is specific.
GATA3 is done, and should be good to order, if Prof Lou want to change any thing.
Regards,
Dawyang
************
chengping wang2 <wangcp@ntu.edu.tw>
To: SuFang Lin <sflin1@gmail.com>
Cc: PJ Lou Alex <pjlou@ntu.edu.tw>, Dawyang Hwang <910208@kmuh.org.tw>, 林玫君 <mayandjean@gmail.com>, 陳贈成 <kmu8501094@yahoo.com.tw>
Sure. I will send different hpv subtypes for this test CP
************
PeiJen, Alex, Lou <pjlou@ntu.edu.tw> Fri, Apr 17, 2015 at 7:35 AM To: SuFang Lin <sflin1@gmail.com>, Dawyang Hwang <910208@kmuh.org.tw>
Cc: ‘王成平 <wangcp@ntu.edu.tw>, 陳贈成 <kmu8501094@yahoo.com.tw>, 林玫君 <mayandjean@gmail.com>
Dear All,
Thanks Dr Huang for your great efforts. We will send the DNA to you, if possible, next week (our DNA quantification reader is out of order and is waiting for being fixed). The GATA3 is a gene of special interest in our lab, so I put it in the list.
All the best,
PeiJen, Alex, Lou MD, PhD
Professor
Department of Otolaryngology
National Taiwan University Hospital and National Taiwan University College of Medicine
************
________

#4由 sufang 在 三, 05/13/2015 – 12:43 發表。
2015/01/16 Daw-yang Hwang Jan 16 Reply to me
CDKN2A (p16) PCR primer
CDKN2A Exon1A_1 CTCTGGTGCCAAAGGGC TCAGTAGCATCAGCACGAGG 210
CDKN2A Exon1A_2 GCGAGTGAGGGTTTTCGTG GCTAGAGACGAATTATCTGTTTACG 226
CDKN2A Exon1B_1 AAGAGGAGGGGCTGGCT CCGTAACTATTCGGTGCGTT 233
CDKN2A Exon1B_2 AGCCTTCGGCTGACTGG ACTTCGTCCTCCAGAGTCGC 180
CDKN2A Exon1C_2 GGGTCGGGTAGAGGAGGTG AACCCCTTCTGAAAACTCCC 223
CDKN2A Exon1C_3 CGACTCTGGAGGACGAAGTT CCCCTTCAGATCTTCTCAGC 211
CDKN2A Exon2_1 ACACAAGCTTCCTTTCCGTC AGCTCCTCAGCCAGGTCC 278
CDKN2A Exon2_2 AGCTGCTGCTGCTCCAC GCATGGTTACTGCCTCTGGT 247
CDKN2A Exon2_3 ACACGCTGGTGGTGCTG GCTTTGGAAGCTCTCAGGGT 230
CDKN2A Exon3 CAGAAGCCAGAGCACATGAA AGGCTCTGGCGCTCCTC 129
CDKN2A Exon3B TGTGCCACACATCTTTGACC CCTGTAGGACCTTCGGTGAC 139
2015/1/22 (Thu) Primers arrived
Templates: OEC-M1 gDNA and TW2.6 gDNA
Primers: Exon3 and Exon3B (從最後兩對測試起)
For a 50-ul reaction:
template 25 ng
F-primer 2 uM
R-primer 2 uM
dNTPs 200 uM
DMSO 5%
10XPCR buffer+MgCl2 5-ul
ddH2O to 50-ul
Touchdown PCR condition (黃醫師的好像還沒有寄過來 我們先用以前做FGFR3-TACC3時用過的,機器上可能還有留底喔)
TD: 65C-55C (SFL Notebook 4, p108)
step 1 94C 2 min
step 2 68C 5 min
step 3 94C 20s
step 4 65C 20s – 0.5C/cycle
step 5 72C 45s
step 6 go to step 3 x 23
step 7 94C 20s
step 8 55C 20s
step 9 72C 45s
step10 go to step 7 x 10
step 11 72C 5min.
End
取 10-ul run on an 1.5% agarose gel.
Break a leg!
2015/02/03 Preliminary results regarding p16 PCR-Seq
Su-Fang Lin <sflin1@gmail.com> to Alex,, 黃道揚醫師, 英潔YCK
Dear Dr. Lou,
我們把p16 PCR的條件試出來了! Primer sequences 部分是由高醫黃道揚醫師提供. 因爲他與我們所上另一個collaborative project的cancer gene list恰好有CDKN2A, 所以就取他已設計好的11對primer來做測試,結果請看下圖:

一剛開始我們拿Exon 3B在OEC-M1與TW2.6 gDNA上做測試.試了幾個條件後、開始懷疑TW2.6細胞中,CDKN2A是否一整段都delete掉了,因此我們加入oral keratinocyte gDNA做另一個control. 上圖右手邊就是這個測試結果 (兩個TW2.6 lane代表不同人抽的DNA), 很顯然TW2.6的CDKN2A locus可能掉了. 我不知道上一次你提到即便IHC+ve的病人DNA做不出PCR, 是否也是這個關係..
以OEC-M1當template, 測試所有11對primer, 目前看來就是在Exon1B_2的地方因long/high GC stretch之故,PCR amplify不出來,其它的amplicon大小都如預期,目前正在Sanger-seq中.. (Exon3 and 3B結果已回來、確認無誤)
這部份先跟您報告到這邊.
Dear 黃醫師, 如果我的primer資訊有誤, 麻煩您直接更正!! 謝謝.
Best,
Su-Fang
2015/02/03 Su-Fang Lin <sflin1@gmail.com> Feb 3 Reply to Alex,, 黃道揚醫師, 英潔YCK, 嬿如YJC
對不起、剛剛那封信的標題裏, results拼錯了…
這一封email主要是try to convince婁醫師,那些珍貴的FFPE DNA,除decode p14/p16 sequence以外,是不是有興趣加看其它如NOTCH1,PIK3CA, MLL2等國外HNSCC mutation marker paper 整理出來的hot genes (2013這篇和2015這一篇)
我和黃醫師討論過, 每50 ng的gDNA應該可以做10–20個全基因掃描. 這裏說的和行動基因/喜來登那一場閻紫宸教授講的東西很像,只是兩邊用的platform不同.我因為想做OSCC FGFR3-TACC3 gene fusion的project而與黃醫師開始有了合作,漸漸地、也開始學到用這套系統做genomic mutations其實快、準而且比傳統PCR-Sanger Seq便宜許多!!
另一個好處是黃醫師會幫忙設計我們想要的基因primer, 我答應他最近要把priority gene list給他, 到時我會cc一份給婁醫師,若你有特別想看的東西可以加上去.
對了,上回我問您說 楊校長/余松良老師那邊的Fluidigm系統可以商借使用嗎? 他們有類似的服務嗎? 還有就是IRB問題,當初這些檢體有寫成像是可以做”genome-wide cancer association” study這類較廣益的用途嗎? 其實在哪邊做都可以, 我要強調的是把這300個珍貴檢體做最大的利用,把我們自己國人的oral cancer, HPV status, p16 sequence, NOTCH1/PIK3CA/EP300 mutation等事情弄清楚, 是一件很有意義的事!
以上!
2015/02/03 Pei-Jen, Alex, Lou Feb 3 Reply to me, 黃道揚醫師
Dear Dr Lin and Dr Huang,
1. 沒問題,我們非常樂意跟妳和黃醫師一起合作!
2. 3百多個口咽癌(oropharyngeal cancer)的DNA已經ready,只要methodology ready就可以on board
3. 如果口咽癌進行順利,接下來可以針對口腔癌(oral cavity)和下咽癌(hypopharyngeal cancer)來作研究
4. IRB沒問題
5. 楊校長的fluidigm也尚在研發階段,沒有對外服務
6. 現在主要的問題是要挑選有興趣的gene!妳先survey一下,我這邊也會看一下,除了e-mail聯絡外,或許過完年找個時間請妳跟黃醫師一起大家開個會直接討論實際的執行方向與細節。
All the best,
Pei-Jen, Alex, Lou MD, PhD
Professor
Department of Otolaryngology
National Taiwan University Hospital and National Taiwan University College of Medicine
2015/02/03 Daw-yang Hwang Feb 3 Reply to Alex,, me, 黃道揚醫師
Dear Dr. Lou and Su-fang,
It is nice to hear that we can collaborated in the oropharyngeal CA. The methodology is ready, please see these 2 publications:
1. http://www.ncbi.nlm.nih.gov/pubmed/24700879
2. http://www.ncbi.nlm.nih.gov/pubmed/24429398
We can meet at discuss the details at your convenience,
Daw-yang
Daw-yang Hwang M.D., Ph.D.
Division of Nephrology
Kaohsiung Medical University
Kaohsiung, Taiwan
On Tue, 3 Feb 2015 10:34:16 +0000, Pei-Jen, Alex, Lou wrote
> Dear Dr Lin and Dr Huang,
>
> 1. 沒問題,我們非常樂意跟妳和黃醫師一起合作!
> 2. 3百多個口咽癌(oropharyngeal cancer)的DNA已經ready,只要methodology ready就可以on board
> 3. 如果口咽癌進行順利,接下來可以針對口腔癌(oral cavity)和下咽癌(hypopharyngeal cancer)來作研究
> 4. IRB沒問題
2015/02/06 Su-Fang Lin <sflin1@gmail.com> Feb 6 Reply to Daw-yang, Alex,, 黃道揚醫師
Dear 黃醫師:
我來交卷了 —> HNSCC的部分.
EGFR mutation在頭頸癌似乎不是主因、主因多為amplification. 所以我只填入亞洲人在lung adenocarcinoma常見的兩個hot spot做分析.
HPV16 和 HPV18 婁醫師已有genotyping data. 若還有空位的話 我想一併以此法測試. 附上去的primer sequences是我們在detect cell line中有無HPV感染時用的序列. 後來想說不一定fit你的rule, 所以還是找出NCBI上原始E6, E7基因片段勞您設計!
謝謝您!
Attachments area attachment HNSCC_SFL.xlsx
2015/02/06 Daw-yang Hwang Feb 6 Reply to me, Alex,
Dear Su-Fang and Dr. Lou,
I will design these PCR primers and will get back to you next week.
Kindly regards,
Daw-yang
2015/02/06 Su-Fang Lin <sflin1@gmail.com> Feb 6 Reply to Daw-yang, Alex,
Dear Daw-yang,
May I remove KDM6A from the list? The more I read, the more I feel it is not worth it. 抱歉!!
2015/02/06 Daw-yang Hwang Feb 6 Reply to me, Alex,
I finished the initail primer design (-KDM6A) on your list, the total number will be around 500. I need to fine tune some of the primers. Let me know if you want to add or delete more genes.
Daw-yang
2015/02/07 Su-Fang Lin <sflin1@gmail.com> Feb 7 Reply to Daw-yang, Alex,
Dear All: Here comes my final gene priority list. 請不吝斧正! (I also include an excel file for your reference)
Have a good weekend~
attachment Gene Priority List_HNSC_SFL.xlsx
2015/02/08 Daw-yang Hwang Feb 8 Reply to me, Alex,
I will wait for prof. Lou’s thinking. We can even decide in the early March after we meet.
Daw-yang
2015/02/09 Pei-Jen, Alex, Lou Feb 9 Reply to Daw-yang, me
Dear Dr Lin and Huang,
1. Thank you so much for Su-Fang’s thorough literature review and prompt reply, and also for Dr Huang’s primer design. I am very keen and look forward to our future collaboration. I’m wondering if we could sit together to discuss practically how to do in detail after lunar new year’s holiday.
2. Our group has a regular meeting on 3/3 evening 5pm at the meeting room of Department of Otolaryngology, National Taiwan University Hospital (New Building, 6F). It’s a Tuesday, so I’m not sure if you are able to make it. If this time does not work for you, then would you be available on 3/7 (Sat) or 3/8 (Sun)?
(Dr. Huang, and Dr Lin if you’re taking the high speed rail as well, please keep the high speed rail ticket and I’ll try to reimburse you the fare.)
3. We have some preliminary results on the association of GATA3 expression (IHC) and the prognosis of head and neck cancer patients. We found that over-expression of GATA3 significantly correlates with patients’ lymph node metastasis, distant metastasis, loco-regional recurrence, disease free survival, and overall survival. From the TCGA database, only 2% head and neck cancer patients harbor GATA3 mutation. From our data, larynx and hypopharyngeal cancer tumors tend to express higher levels of GATA3 while oropharynx and oral cavity cancer tumors tend to express lower levels of GATA3. Since the role of GATA3 in the carcinogenesis and prognosis of head and neck cancer has not been elucidated, I’m wondering if we should add GATA3 in the list of our fluidigm analyses? Please give me your thoughts and comments!
All the best,
Pei-Jen, Alex, Lou MD, PhD
2015/02/09 Daw-yang Hwang Feb 9 Reply to Alex,, me
Dear Prof. Lou and Su-Fang,
3/3 is fine for me and I can arrive NTU before 5PM. There is no problem to include GATA3 in the Fluidigm analysis. I believe you and Su-fang have more feeling in what genes to be selected, and I can provide all necessary technical support.
Warmly regards,
Daw-yang
2015/02/09 Su-Fang Lin <sflin1@gmail.com> Reply to Daw-yang, Alex,
Dear Drs. Lou and Hwang,
Sorry for the belated reply. I was hosting Drs. 陳華鍵 and 陳淑貞 from 行動基因 whole day today. You may not know that both of them worked for NHRI IBPR (生技藥研所) before. Today they 回娘家給演講!
3/3 (二) 我沒問題. 我們就醬子說定囉!
LINE的話我和黃醫師已有”群組”. 我把我的QR code給婁醫師 (見下附檔). 請您把我掃進去. 然後我幫您加進來! This is a good toy for communication.
Best,
Su-Fang
2015/02/10 Pei-Jen, Alex, Lou Feb 10 Reply to me, Daw-yang
Dear All,
Great! Let’s make it at 5pm 3/3 (Tue), 臺大醫院新大樓1樓Lobby,I’ll meet you up there and escort you to our meeting room. Could it be possible that Dr Huang prepare a couple of slides to briefly introduce the fluidigm system to my young colleagues and Dr Lin briefly introduce how you select the genes of interest? This is a group meeting and only me and 2 younger PI in our department will join us for discussion. So be relax and casual! Look forward to seeing you soon. Happy new year!
All the best,
Pei-Jen, Alex, Lou MD, PhD
Professor
Department of Otolaryngology
National Taiwan University Hospital and National Taiwan University College of Medicine
2015/02/10 Daw-yang Hwang Feb 10 Reply to Alex,, me
Dear Prof. Lou,
I can prepare few slides about the Fluidigm Access Array system on 3/3. See you there!
Warmly regards,
Daw-yang
On Mon, 9 Feb 2015 23:47:25 +0000, Pei-Jen, Alex, Lou wrote
Daw-yang Hwang M.D., Ph.D.
Division of Nephrology
Kaohsiung Medical University
Kaohsiung, Taiwan
———————————
2015/02/11 LINE Posts
Su-Fang:
–我們所上oral cancer team 在二期癌症計畫的「缺口計畫」裡propose study “oral potentially malignant disorders”. 拿了一個1300萬的project. 這些錢主要用來「買檢體」合作醫院包括 中榮、北榮、成大、高醫、中山、中國醫大.
–OPMD (口腔癌前病變) 的檢體 (archived 或是 prospective) 有個很難做的地方就是小、而且聽說為了切乾淨不得不夾雜許多正常細胞. 我個人寫的子計劃是檢查葉酸代謝相關酵素 MTHFR 兩個functional SNPs在 100例 OPMD patients中的 pattern. 接下來我會 由此延伸至folate deficiency -mediated genomewide hypomethylation/focal hypermethylation. 最終目標是找出 methyated DNAs that are capable of identifying OPMD with high-risk in malignant transformation.
–那天聽婁醫師說 你們那邊也有人做OPMD, 不曉得你們團隊的研究興趣主要是在哪邊? 有沒有機會可以相互切磋一下, 尤其是 genetic and epigenetic factors. 我覺得病程後面OSCC部分國內外已經做很多了, 而且現在又有像道揚這種敏感度高的NGS技術, 所以拿OSCC的data來模擬從OPMD惡化成OSCC的主要因子會是哪些, 然後把high-risk OPMD 病人篩出來 應是極為可行的. (以上)
–Daw-yang, 趁你在線上問一個問題, 那天ACT的華鍵、淑貞博士說 copy number variation (CNV) 測試他們會用到一些normal control當baseline. 那你這邊是說limited PCR, 除此之外, 還需normal controls嗎? thank you!
Daw-Yang:
–There is no really NORMAL CONTROL, I think they use some healthy individual as normal control. Or you have to do some aCGH, real-time PCR as you control. I know some company sell these normal control samples
–Just like real time pcr, people use TERT gene as internal normal control
Alex:
–Our oral pre-cancer study is a collaborative prospective study with the NCI/NIH USA. This ia a 7-year project. So far we only collect patient samples (including blood, cytobrush, biopsy specimen and epidemiological data). Maybe we can talk a bit on the future collaboration in our 3/3 meeting on what to do on our collected samples. SuFang is right that the biopsy samples are very small and difficult to get large amount of DNA or RNA for further study
–In our Biosignature study this year, we propose to perform detailed genome wide screening (exome, RNA-seq and RRBS) on paired normal, pre-cancer and cancer tissues of 10 OSCC patients with different environmental exposures as a first tier screening for mutations, epimutaions and differentially expressed genes.
–We propose to perform whole exome sequencing, mRNA sequencing (RNA-seq) and reduced representation bisulfite sequencing (RRBS).
2015/03/03 First meeting at NTUH
–七年3000例, 弄清楚malignant transformation rate. 但目前還沒有規劃要做什麼, 只是先做苦工…
–Alan Hildesheim 艾倫·希爾德斯海姆 http://dceg.cancer.gov/about/staff-directory/biographies/A-J/hildesheim-allan
–查阿牛 Anil Chaturvedi 下個月 (四月) 會來! http://dceg.cancer.gov/about/staff-directory/biographies/A-J/chaturvedi-anil
|
|
|
|
|
|
|
|
|
#5由 sufang 在 日, 03/01/2015 – 13:54 發表。
2015/01/12
Dear 婁教授,
您好, 很冒昧直接寫信給您. 我叫林素芳, 昨天早上在喜來登飯店B2F和您請教過CDKN2A/ p16與oral cancer關聯的那一個! Here is my brief CV on the web. 口腔癌是國衛院癌研所二個重點研究癌症之一。另外去年年底,在二期癌症缺口計畫一項,癌研所亦獲得補助整合中南部六家醫院口腔癌前病變(OPMD, oral potentially malignant disorders)檢體之收集與高危險群之篩檢及惡性轉化之預防.
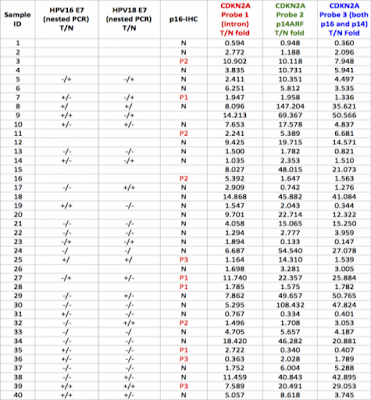
之前我在核對所裡microarray database(Illumina platform)時,發現台灣OSCC檢體(n=40) CDKN2A這個loci的transcript, tumor : normal是不降反升; 同事拿microarray result 和同時做的methylome array相比,也發現methylation程度和transcript量竟然成正比的怪現象; 我也請成大蕭振仁醫師幫忙做同一批檢體的p16 IHC staining 或是以PCR測試HPV16/18之E6/E7, 但結果還是兜不起來..
昨日聽到您的一些p16 mutation結果真是興奮極了! 不幸, 回來與同事們分享時卻又落東落西的,說不出一個完整故事..嗚嗚嗚..因此想說是否有幸能邀請您到敝所來給一個演講,介紹您在頭頸癌方面的研究成果!
我們所分竹南、台南兩個地方 您可能碰到過的蕭振仁醫師、張書銘老師是在台南,竹南這邊大多做basic science, 台南那邊多是臨床醫師 (including陳立宗所長)。每週一13:30-15:00是所裡固定Departmental Seminar時間, 採視訊方式。婁醫師如果您的星期一下午不方便的話,其它weekday我們也可以配合。從台北來的外賓我們會派車到竹北高鐵站接送,約30 min車程可達院區。中午與竹南這邊的PI用個簡餐、聊聊天,然後就是talk. 另外會有一份微薄的演講費.
Summary:
Time: Preferably Mondays 13:30-15:00 or your convenient days.
Place: NHRI 竹南院區
Title: Whatever you want! Hint: p16/CDKN2A in oral cancer, Taiwan biobank
如果您可以撥出時間來一趟的話,我再請秘書把目前空著的slot裏出來,寄回給您挑選可行之日.
Again, apologize for my rudeness.
Best,
Su-Fang
2015/01/13
Pei-Jen, Alex, Lou to me
Dear Prof Lin,
Have you done the sequencing of p16 on these sample?
Pei-Jen, Alex, Lou MD, PhD
Professor
Department of Otolaryngology
National Taiwan University Hospital and National Taiwan University College of Medicine
Su-Fang Lin to Alex 2015/01/13
Nope, we only have expression array data for the 40 tumor/normal paired samples.
However, I do have RNA-Seq data for several Taiwanese OSCC cell lines (TW2.6, OEC-M1, OC3 and couple more from 長庚鄭恩加老師).
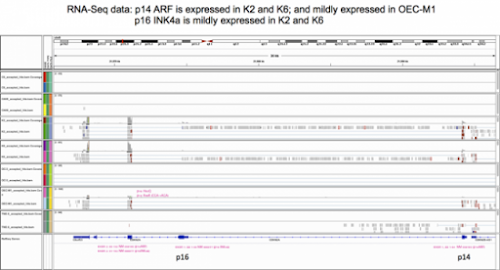
剛剛快速掃了一下, OSCC cell line中只有OEC-M1的CDKN2A locus有reads. 其中比較顯著的mutation是p16 (NM_000077) H83H, CAC->CAT, 同一個位置在
p14 (NM_058195) R98, CGA ->TGA (stop codon!!) 咦? 那不就斷了…
另外 p14 I49, Q50那邊好像也有一點non-synomous mutation. 不過read不高. 不是很確定.
您會建議我們在病人tissue DNA上做PCR-sequencing嗎?
Pei-Jen, Alex, Lou Jan 13 Reply to me
Dear Prof Lin,
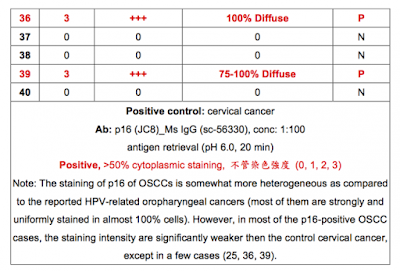
1. p16在頭頸癌的故事可能非常複雜,在國外的研究發現p16可能是”頭頸癌”最常見的突變基因。然而頭頸癌包括鼻咽,口腔,口咽,下咽和喉癌等,國內”口腔癌”的研究發現p16(IHC)常會over-express且correlates with poor survival。國外”口咽癌”的研究發現p16 (IHC) correlates with HPV (ISH or genotyping),patients with p16 (+) have better treatment outcome to chemoradiotherapy and survival.
2. 我個人認為,OSCC中IHC 染到的p16可能是mutant form而國外口咽癌中IHC染到的p16可能是wild type。
3. 我們最近check了3百多個國人口咽癌的tissue sample, 每個都作了p16 IHC and HPV genotyping,所有病人tumor的DNA都已從paraffin block抽出來了,接下來想作p16 PCR sequencing,如果你有興趣可以跟我們合作,PCR-sequencing的部分給你作,相信一定會有very interesting findings.
4. 事實上p16 PCR-sequencing seems not to be very easy. 我們先前test了幾個sample,不過好像不太作得出來(even in p16 IHC (+) cases),你可以先用cell line試試,如果實驗條件穩定,我可以把tumor DNA送去給你作。
5. for your 40 OSCC samples, I’ll suggest to do p16 PCR-sequencing. This will clarified many unanswered questions.
Pei-Jen, Alex, Lou MD, PhD
Professor
Department of Otolaryngology
National Taiwan University Hospital and National Taiwan University College of Medicine
Su-Fang Lin <sflin1@gmail.com> Jan 13 Reply to Alex,
好耶 我很有興趣幫忙做p16 PCR-sequencing! 謝謝你的寶貴information, it does make sense to me that high IHC staining indeed was due to mutant p16. 我明天到office把相關資料查清楚之後再和你做進一步報告.
Su-Fang Lin <sflin1@gmail.com> AttachmentsJan 14 Reply to Alex,, bcc: 黃道揚
Dear Dr. Lou:
Here is my little research regarding CDKN2A:
(slide 1) CDKN2A locus encodes two proteins: p14 and p16. 兩個蛋白的exon 1不同. 國衛院使用的Illumina microarray platform有三個probe針對這個locus, 不幸地, probe 1 落在intron裏, probe 2 is good for p14, probe 3 落在兩者的3UTR. 因此,嚴格講來我們那40對檢體的array data並沒有辦法確切指出誰的p16 transcript是高的.
(slide 2) 詳述p14的3個exon及translated protein. 這裏有兩點值得一提: (1) Sigma 有一支抗體是用紫色label的68個aa當抗原,因此是在染p14 請小心你們染IHC時不是用這支抗體. 因為p14和anti-apoptosis有關-> strong staining correlates with poor survival -> make sense. (2)前天我說RNA-Seq data顯示OEC-M1 cell line在C452T這個mutation, 將造成p16提前斷掉.
(slide 3) 詳述p16的3個exon及translated protein. 你可以看到雖然p14和p16共用exon 2 and 3, 但是frameshift之故,剛剛造成stop codon的位點在p16是一個silent mutation (C555T/H83H)
(slide 4) 強迫做p14及p16的 aa seq alignment, 並未看出特別的conserved region. (我原來懷疑兩者有share一些
抗原性,混淆IHC的結果)
Now, back to p16 PCR-sequencing的實驗. 婁醫師,您有聽過Fluidigm Access Array的系統嗎? (eg from 楊校長實驗室家的人) 簡單地說就是combine PCR + NGS 的技術,可以很快、極有效率地做targeted re-sequencing的實驗.
昨天你問我是不是有興趣 resequencing p16, 我就一直在想是不是該跟你推薦做這個. 我也最近因為認識高醫腎臟科黃道揚醫師 而正在學習相關技術.以50 ng的FFPE DNA而言、利用這套系統可以同時看包含p16在內的許多基因的mutation (對了, 那天喜來登飯店ACT GENOMICS發的DM中的p4, 原理非常相似. 惟黃醫師自己設計primer與barcode, cost cutdown很多)
Summary for the time being:
1. Let’s PCR-sequencing p16 for those 300 samples!
2. 用DNA當template的話沒有道理PCR不出來啊, 請給我一點時間找些文獻 看看是怎麼一回事.
先降子..Good Night.
2015/1/19 晚間LINE 對話
SFL: Dear DW, 設計primer真不是一件簡單的事.我拿著你給的sequence照著要去confirm看位置對不對,花了一個早上還搞不定. 後來下午好不容易把exon1的部份弄完後,就跟助理說, 訂吧, 一定對的, 我眼睛好酸啦(scream)
SFL: 現在想請教你: primer 回來後,check PCR的條件. For a 10-ul reaction: template 50 ng, dNTP 200 microM each, MgCl2 4.5mM, 5% DMSO. Annealing 60C, ~ 35 cycles. OK 嗎? 感謝!!
DWH: Dear Sufang, I will use only 5ng DNA. Any protocol you use should be ok. I usually use master PCR mixture to save time. I never test the Tm of my primer. I only use touch-down protocol.
DWH: If you are interesting in my touch-down protocol, I can ask my assistant mail to your assistant.
DWH: The CDKN2A has 3 splicing forms differ in the first exon, so I make them as 1A, 1B, 1C. This is the tedious part for some genes.
DWH: My suggestion for your oral cancer is you can pick some genes so I can prepare them.
|
|
|
|
|
|
|
|
|
#6由 sufang 在 一, 01/19/2015 – 09:59 發表。
1. Int J Cancer. 2014 Oct 4. doi: 10.1002/ijc.29254. [Epub ahead of print] A high and increasing HPV prevalence in tonsillar cancers in Eastern Denmark, 2000-2010: The largest registry-based study to date. Garnaes E1, Kiss K, Andersen L, Therkildsen MH, Franzmann MB, Filtenborg-Barnkob B, Hoegdall E, Krenk L, Josiassen M, Lajer CB, Specht L, Frederiksen K, Friis-Hansen L, Nielsen FC, Kjaer SK, Norrild B, vonBuchwald C.
Abstract
The aim was to explore whether the incidence of tonsillar squamous cell carcinomas (TSCCs) increased in Eastern Denmark, 2000-2010, and whether HPV could explain the increase, and to assess the association of HPV prevalence with gender, age, and origin (i.e., the certainty of tonsillar tumor origin). We applied HPV DNA PCR and p16 immunohistochemistry to all TSCCs registered in the Danish Head and Neck Cancer Group (DAHANCA) and in the Danish Pathology Data Bank (n = 632). Pathologists reviewed and subdivided the tumors into two groups: specified and non-specified TSCCs. Approximately 10% of HPV-positive tumors was genotyped by amplicon next-generation sequencing. The overall crude incidence of TSCCs increased significantly (2.7% per year) and was explained by an increasing incidence of HPV-positive TSCCs (4.9% per year). The overall HPV prevalence was 58%, with HPV16 being the predominant HPV type. In multivariate analysis, the HPV prevalence was associated with age (<55 vs. >60 years) (OR, 1.72; 95% CI 1.13-2.63) and origin (non-specified vs. specified TSCCs) (OR, 0.15; 95% CI 0.11-0.22). The association of HPV prevalence with origin increased over time in specified TSCCs (OR per year, 1.10; 95% CI 1.01-1.19), whereas no change over time was observed among non-specified TSCCs (OR per year, 0.99; 95% CI 0.90-1.08). In conclusion, the observed increase in the number of HPV-positive TSCCs can explain the increasing number of TSCCs in Eastern Denmark, 2000-2010. HPV prevalence was associated with younger patients (<55 years) and a high certainty of tonsillar tumor origin. © 2014 Wiley Periodicals, Inc. PMID: 25283302
7. ONLINE FIRST: HPV Status Found to be Strong Predictor of Survival in Recurrent Oropharyngeal Cancer
Patients with advanced recurrent oropharyngeal squamous cell carcinoma have a better chance of survival if their tumors are positive for human papillomavirus (HPV), according to a new study. HPV-positive status should therefore be a stratification factor for future clinical trials involving patients with recurrent or metastatic disease, the authors of the new findings suggest.
The research, by Johns Hopkins scientists and now available online ahead of print in the Journal of Clinical Oncology (doi: 10.1200/JCO.2014.55.1937), included 181 patients with stages III-IV oropharynx cancer who were enrolled in the Radiation Therapy Oncology Group trials 0129 or 0522 and who had known tumor HPV type p16 status and local, regional, and/or distant progression after receiving platinum-based chemoradiotherapy.
Of the total participants, 105 were p16-positive and 76 were p16-negative. After a median follow-up period of four years after disease progression, patients with HPV p16-positive oropharyngeal cancer had significantly increased survival rates compared with patients who were HPV p16-negative – two-year overall survival rates of approximately 55 and 28 percent, respectively, and median survival times of 2.6 versus 0.8 years, respectively.
|
__________
#7由 sufang 在 日, 03/01/2015 – 17:56 發表。
HPV infection status in 40TN
2013/07/29 from YCK_27組sample

2013/08/09 from Ya-Wen
陳雅雯 <ywc@nhri.org.tw> Fri, Aug 9, 2013 at 9:40 PM To: 林素芳 <sflin@nhri.org.tw>
Just happen to remember that we can detect secreted p16 in cells with p16 overexpression. We have worked with Dr. Huang TS to check the secreted p16 in serum of HCCs. However, finally I quit from the project and I guess that
the project was ended. What I am thinking now is that we can observe up-regulated p16 in 40 pairs of OSCC.
Maybe we can detect p16 from serum of OSCC. Talk to you later. Ya-Wen
2013/08/23
       
2013/08/29
Gmail : Analysis of p16INK4A expression of oral squamous cell carcinomas in Taiwan: Prognostic correlation without relevance to betel quid consumption – Chen – 2012 – Journal of Surgical Oncology – Wiley Online Library
Ya-Wen Chen <980727@nhri.org.tw> Thu, Aug 29, 2013 at 8:30 PM To: Sflin <sflin@nhri.org.tw>
http://onlinelibrary.wiley.com/doi/10.1002/jso.23067/pdf
2013/08/30
NM_000077, VARIANT 1, ENCODING P16 INK4A
Summary: This gene generates several transcript variants which differ in their first exons. At least three alternatively spliced variants encoding distinct proteins have been reported, two of which encode structurally related isoforms known to function as inhibitors of CDK4 kinase. The remaining transcript includes an alternate first exon located 20 Kb upstream of the remainder of the gene; this transcript contains an alternate open reading frame (ARF) that specifies a protein which is structurally unrelated to the products of the other variants. This ARF product functions as a stabilizer of the tumor suppressor protein p53 as it can interact with, and sequester, the E3 ubiquitin-protein ligase MDM2, a protein responsible for the degradation of p53. In spite of the structural and functional differences, the CDK inhibitor isoforms and the ARF product encoded by this gene, through the regulatory roles of CDK4 and p53 in cell cycle G1 progression, share a common functionality in cell cycle G1 control. This gene is frequently mutated or deleted in a wide variety of tumors, and is known to be an important tumor suppressor gene. [provided by RefSeq, Sep 2012].
2013/09/04
Gamil: Fw: QuantiVirus® HPV E6/E7 mRNA Test for Cervical and Head-Neck Cancer
Ya-Wen Chen <980727@nhri.org.tw> To: Sflin <sflin@nhri.org.tw>
HPV
test
Ya-Wen
Wed, Sep 4, 2013 at 8:10 AM
2013/09/10
2013/10/11
2014/Feb-Mar
|
|